WP5 – Identification and validation of novel therapeutic targets
Investigators and institutions: Krister Wennerberg, BRIC, PI; Jérémy Ariey-Bonnet, BRIC, postdoc)
Rationale
Glioblastoma (GBM) growth and relapse is thought to be driven by a small subpopulation of glioblastoma stem cells (GSCs) that display self-renewal and tumor-initiating capacities. Importantly, following surgery that removes the tumor bulk, and adjuvant chemo-radio therapy, this population can be reactivated and drive relapse. In this context, there is a need to develop approaches aiming at understanding and targeting the biology mechanisms driving survival and self-renewal of this cell subpopulation.
In this WP, we will identify proteins as being required for the maintenance of the stem-like cell population in GBM and qualify them as therapeutic targets for the development of precision medicine in GBM.
Objectives
- Identification of putative genes required for GBM maintenance from public CRISPR-based functional genetic screening data.
- Detailed analyses of genetic dependencies in GBM and how they can be turned into therapeutic approaches.
- Preclinical validation of new therapeutic strategies in GBM and how these could be stratified.
Approach
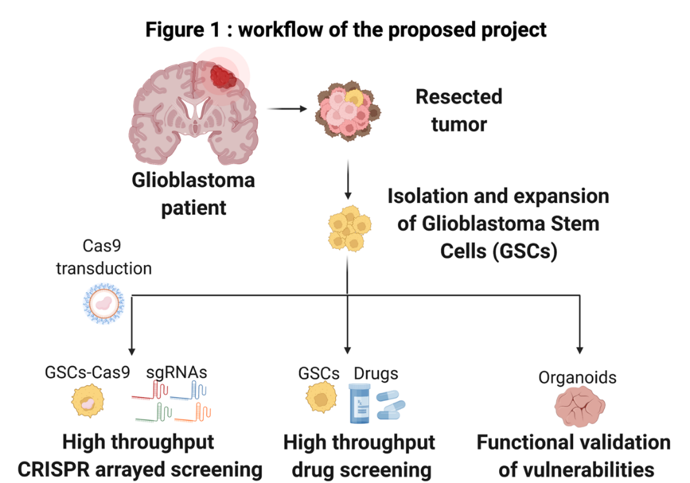
GSC cultures growing as neurospheres mirror the genotype of primary tumors. This model can readily be established from patient samples and preserve the stem cell properties of GBM cells.
Using a protocol established in the lab, we will dissociate primary tumor samples from 10 GBM patients (2-3 from each of the 4 major GBM subtypes; proneural, neural, classical or mesenchymal subtype) and establish neurosphere cultures.
We will then generate Cas9-expressing neurosphere models using a lentiCas9 blast virus from our lab. These Cas9-expressing neurosphere culture variants will be transfected with a customized sgRNA library targeting putative genes involved in GSCs survival.
To design the sgRNA library that we will screen, we analyzed published genome-wide pooled CRISPR screens (Toledo et al. Cell Rep. 2015; MacLeod et al, Cell Rep. 2019) of 12 GSC cultures. These analyses led us to the identification of 1654 putative GSC-specific gene dependencies. This gene set represents genes that appear essential for growth in the published GSC cultures, but that are not general essential genes in broader sets of cell models. Among the genes are multiple well-known GBM/GSC gene dependencies (such as EGFR, AKT1, PIK3CA), arguing for the relevance of the approach. However, most identified genetic vulnerabilities have not previously been explored as therapeutic targets in GBM.
The dependency on the 1654 identified genes will be tested one-by-one in the Cas9-expressing GSC cultures using high throughput CRISPR arrayed screening. Testing will be done in 384-well format using transfectable guide RNAs. This will uniquely allow for a high throughput phenotypic evaluation of each gene knockout. The impact of the knockouts will be assessed on the Cas9-expressing GSC cultures in organoid-type growth conditions where the cells are embedded in 3-D extracellular matrix, which induces a greater intra-culture differentiational heterogeneity. Overall viability will be determined using CellTiter-Glo assay and cell subpopulation drug responses will be assessed by immunofluorescence markers using our high content microscope. One week after knockout, the organoids will be fixed and stained for GSC-like cells and differentiated cells within the organoids.
A key goal of the project is to convert the gene dependencies into therapeutic opportunities. To do this, we are searching for drugs and drug candidates that directly inhibit the function of the proteins the dependency genes code for. However, we know that there are only selective inhibitors for a small percentage of all gene products. Therefore, we are also analyzing biochemical pathways and processes that are enriched with dependency hits with the expectation that these pathways/processes can be relevant drug targets. Analyses of dependency-enriched pathways and drugs that target them is ongoing. Both known GBM pathways (such as FGFR, EGFR, PI3K-AKT, stem cell, DNA repair pathways) and previously non-explored pathways have been identified. In the end, we expect to build a collection of about 300 compounds to be tested on the GSC-derived organoid cultures in the same way as the CRISPR-driven gene knockout studies. A focus will be on drugs inhibiting targets that are not explored in WP2 as well as small molecules that can penetrate (or are predicted to penetrate) the blood-brain barrier. Overall viability and cell subtype drug responses will be assessed by high throughput microscopy to identify individualized drug response “fingerprints” of each patient sample and a phenotypic stratification across the tested cohort. The cell subtype drug response analysis will allow us to identify drugs that target different phenotypic subpopulations, including GSCs.
Drug responses and gene knockout experiments from chemical and genetic screens will be used to build a confirmed dependency map in the different GBM models and this information will be compared to genetic information from WP1 to make links between somatic genetics and the novel therapeutic GBM vulnerabilities. Top hits, as single agents and in combination with temozolomide, will be validated in long-term in vitro cultures (up to two months) to identify agents that fully eradicate the GBM cells using the organoid model we established in the lab. Mechanism of action of long-term effective hits will be explored and one or two top-validating agents will finally be tested in orthotopic xenograft animal models developed in WP3.
Endpoints
Identification of new druggable GSC-directed molecular targets.
Mechanism of action of targeting GSCs
Preclinical validation of the targets and drugs inhibiting them.
Expected impact
Preclinical data supporting potential clinical exploration of new GSC-targeting drugs.